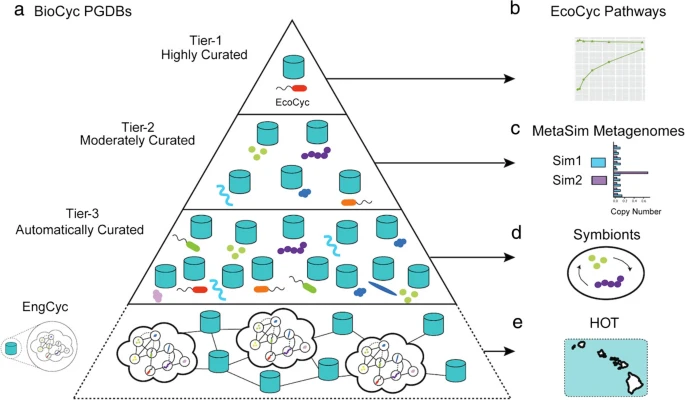
Global Scale Metabolic Pathway Reconstruction From Environmental
Genomes
For more than 3.5 billion years, microorganisms have been the
dominant form of life on Earth, mediating global fluxes of matter
and energy. Over the past decade, high-throughput sequencing and
mass spectrometry platforms generating multi-omic sequence
information (DNA, RNA, protein and metabolites) which contain
information about the function and identity of microbial life,
have transformed our perception of this microcosmos, illuminating
microbial dark matter and conceptually linking microorganisms at
the individual, population, and community levels to a wide range
of ecosystem functions and services. Despite the power and promise
of this new perception, a persistent paucity of scalable software
tools to mine, monitor, and interact with environmental sequence
information limits knowledge creation and translation. This is
especially vexing in a time of climate change when microbial
community metabolism offers a virtual blueprint to rebuild our
global future in more sustainable ways. EngCyc is the next step in
overcoming this challenge.